Example: Likelihood fitting a Bivariate Gaussian¶
In this example, we shall perform likelihood fitting to a bivariate normal distribution, to demonstrate how symfit’s API can easily be used to perform likelihood fitting on multivariate problems.
In this example, we sample from a bivariate normal distribution with a significant correlation of \(\rho = 0.6\) between \(x\) and \(y\). We see that this is extracted from the data relatively straightforwardly.
[1]:
import numpy as np
from symfit import Variable, Parameter, Fit
from symfit.core.objectives import LogLikelihood
from symfit.distributions import BivariateGaussian
import matplotlib.pyplot as plt
Build a model corresponding to a bivariate normal distribution.
[2]:
x = Variable('x')
y = Variable('y')
x0 = Parameter('x0', value=0.6, min=0.5, max=0.7)
sig_x = Parameter('sig_x', value=0.1, max=1.0)
y0 = Parameter('y0', value=0.7, min=0.6, max=0.9)
sig_y = Parameter('sig_y', value=0.05, max=1.0)
rho = Parameter('rho', value=0.001, min=-1, max=1)
pdf = BivariateGaussian(x=x, mu_x=x0, sig_x=sig_x, y=y, mu_y=y0,
sig_y=sig_y, rho=rho)
Generate mock data
[3]:
# Draw 100000 samples from a bivariate distribution
mean = [0.59, 0.8]
corr = 0.6
cov = np.array([[0.11 ** 2, 0.11 * 0.23 * corr],
[0.11 * 0.23 * corr, 0.23 ** 2]])
np.random.seed(42)
xdata, ydata = np.random.multivariate_normal(mean, cov, 100000).T
[4]:
hist = plt.hist2d(xdata, ydata, bins=(100, 100))
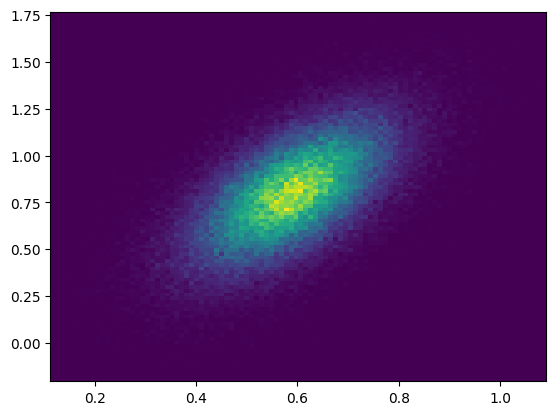
Finally, we perform the fit to this mock data using the LogLikelihood objective.
[5]:
fit = Fit(pdf, x=xdata, y=ydata, objective=LogLikelihood)
fit_result = fit.execute()
print(fit_result)
/home/docs/checkouts/readthedocs.org/user_builds/symfit/envs/stable/lib/python3.7/site-packages/symfit/core/objectives.py:459: RuntimeWarning: divide by zero encountered in log
[np.nansum(np.log(component)) for component in evaluated_func]
/home/docs/checkouts/readthedocs.org/user_builds/symfit/envs/stable/lib/python3.7/site-packages/symfit/core/objectives.py:459: RuntimeWarning: invalid value encountered in log
[np.nansum(np.log(component)) for component in evaluated_func]
/home/docs/checkouts/readthedocs.org/user_builds/symfit/envs/stable/lib/python3.7/site-packages/symfit/core/objectives.py:485: RuntimeWarning: divide by zero encountered in true_divide
df / component
/home/docs/checkouts/readthedocs.org/user_builds/symfit/envs/stable/lib/python3.7/site-packages/symfit/core/objectives.py:485: RuntimeWarning: invalid value encountered in true_divide
df / component
/home/docs/checkouts/readthedocs.org/user_builds/symfit/envs/stable/lib/python3.7/site-packages/symfit/core/fit_results.py:256: RuntimeWarning: overflow encountered in exp
gof_qualifiers['likelihood'] = np.exp(gof_qualifiers['log_likelihood'])
Parameter Value Standard Deviation
rho 6.026420e-01 2.013810e-03
sig_x 1.100898e-01 2.461684e-04
sig_y 2.303400e-01 5.150556e-04
x0 5.901317e-01 3.481346e-04
y0 8.014040e-01 7.283990e-04
Status message CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH
Number of iterations 22
Objective <symfit.core.objectives.LogLikelihood object at 0x7f980b4bc2d0>
Minimizer <symfit.core.minimizers.LBFGSB object at 0x7f980b486910>
Goodness of fit qualifiers:
likelihood inf
log_likelihood 106241.24669486462
objective_value -106241.24669486462
We see that this result is in agreement with our data.